Sophisticated free app for molecular visualization
iRASPA for macOS employs the latest visualization technologies with stunning performance
27 March 2018
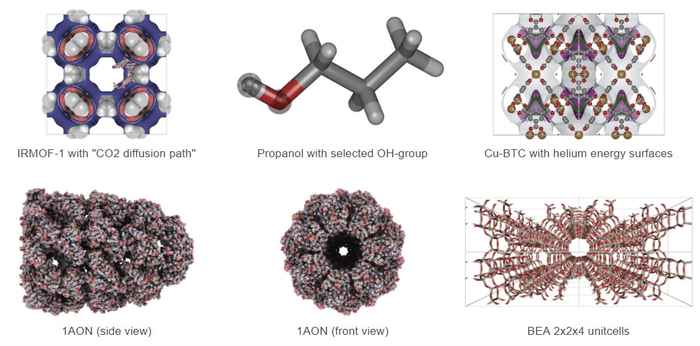
iRASPA is a visualization package with editing capabilities aimed at materials science. Controlled by means of a versatile user interface it produces high quality images of complex molecules, metals, metal-oxides, ceramics, biomaterials, zeolites, clays, and metal-organic frameworks.
iRASPA is exclusively for macOS and as such can leverage the latest visualization technologies with stunning performance. iRASPA extensively utilizes GPU computing. For example, void-fractions and surface areas can be computed in a fraction of a second for small/medium structures and in a few seconds for very large unit cells. It can handle large structures (hundreds of thousands of atoms), including ambient occlusion, with high frame rates.
A user comment in the app store: "A must have for any scientist working with porous materials".
Acces to database
Via iCloud, iRASPA has access to the CoRE Metal-Organic Frameworks database containing approximately 8000 structures. All the structures can be screened (in real-time) using user-defined predicates. The cloud structures can be queried for surface areas, void fraction, and other pore structure properties.
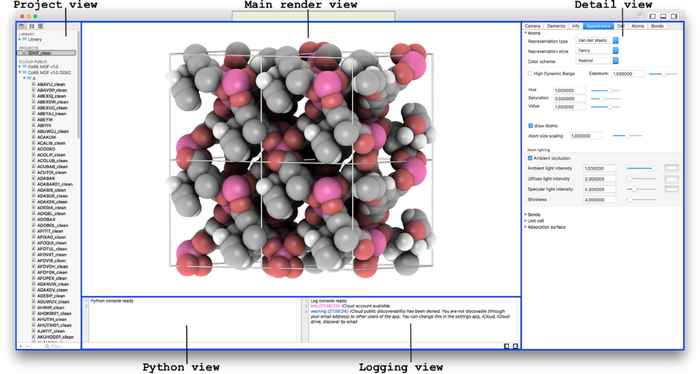
Main features of iRASPA are:
- structure creation and editing;
- creating high-quality pictures and movies;
- ambient occlusion and high-dynamic range rendering;
- collage of structures;
- (transparent) adsorption surfaces;
- text-annotation;
- cell replicas and supercells;
- symmetry operations like space group and primitive cell detection;
- screening of structures using user-defined predicates;
- GPU-computation of void-fraction and surface areas in a matter of seconds.
iRASPA has been created by David Dubbeldam (University of Amsterdam, The Netherlands), Sofia Calero (Universidad Pablo de Olavide, Seville, Spain) and Thijs Vlugt (Delft University of Technology, The Netherlands) with contributions from Randall Q. Snurr (Northwestern University, Evanston, USA).
A detailed description of iRASPA and its use is presented in a recent open-access article in Molecular Simulation:
David Dubbeldam, Sofía Calero & Thijs J.H. Vlugt (2018) iRASPA: GPU-accelerated visualization software for materials scientists, Molecular Simulation, 44:8, 653-676, DOI: 10.1080/08927022.2018.1426855